3.3. Scikit-image: image processing¶
Author: Emmanuelle Gouillart
scikit-image is a Python package dedicated
to image processing, and using natively NumPy arrays as image objects.
This chapter describes how to use scikit-image on various image
processing tasks, and insists on the link with other scientific Python
modules such as NumPy and SciPy.
See also
For basic image manipulation, such as image cropping or simple filtering, a large number of simple operations can be realized with NumPy and SciPy only. See Image manipulation and processing using NumPy and SciPy.
Note that you should be familiar with the content of the previous chapter before reading the current one, as basic operations such as masking and labeling are a prerequisite.
3.3.1. Introduction and concepts¶
Images are NumPy’s arrays np.ndarray
- image:
np.ndarray- pixels:
array values:
a[2, 3]- channels:
array dimensions
- image encoding:
dtype(np.uint8,np.uint16,np.float)- filters:
functions (
numpy,skimage,scipy)
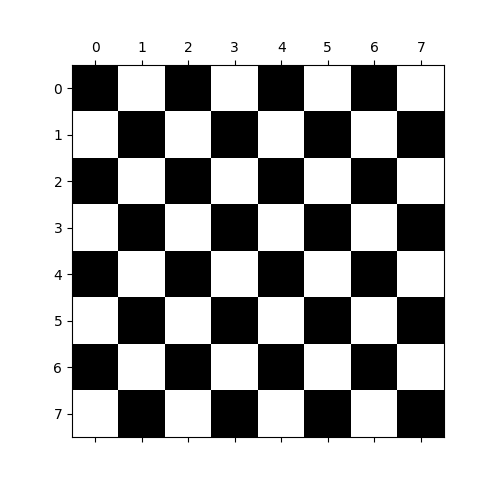
>>> import numpy as np
>>> check = np.zeros((8, 8))
>>> check[::2, 1::2] = 1
>>> check[1::2, ::2] = 1
>>> import matplotlib.pyplot as plt
>>> plt.imshow(check, cmap='gray', interpolation='nearest')
3.3.1.1. scikit-image and the SciPy ecosystem¶
Recent versions of scikit-image is packaged in most scientific Python
distributions, such as Anaconda or Enthought Canopy. It is also packaged
for Ubuntu/Debian.
>>> import skimage
>>> from skimage import data # most functions are in subpackages
Most scikit-image functions take NumPy ndarrays as arguments
>>> camera = data.camera()
>>> camera.dtype
dtype('uint8')
>>> camera.shape
(512, 512)
>>> from skimage import filters
>>> filtered_camera = filters.gaussian(camera, 1)
>>> type(filtered_camera)
<type 'numpy.ndarray'>
Other Python packages are available for image processing and work with NumPy arrays:
scipy.ndimage: for nd-arrays. Basic filtering, mathematical morphology, regions properties
Also, powerful image processing libraries have Python bindings:
(but they are less Pythonic and NumPy friendly, to a variable extent).
3.3.1.2. What’s to be found in scikit-image¶
Website: https://scikit-image.org/
Gallery of examples: https://scikit-image.org/docs/stable/auto_examples/
Different kinds of functions, from boilerplate utility functions to high-level recent algorithms.
Filters: functions transforming images into other images.
NumPy machinery
Common filtering algorithms
Data reduction functions: computation of image histogram, position of local maxima, of corners, etc.
Other actions: I/O, visualization, etc.
3.3.2. Input/output, data types and colorspaces¶
I/O: skimage.io
>>> from skimage import io
Reading from files: skimage.io.imread()
>>> import os
>>> filename = os.path.join(skimage.data_dir, 'camera.png')
>>> camera = io.imread(filename)

Works with all data formats supported by the Python Imaging Library
(or any other I/O plugin provided to imread with the plugin
keyword argument).
Also works with URL image paths:
>>> logo = io.imread('https://scikit-image.org/_static/img/logo.png')
Saving to files:
>>> io.imsave('local_logo.png', logo)
(imsave also uses an external plugin such as PIL)
3.3.2.1. Data types¶

Image ndarrays can be represented either by integers (signed or unsigned) or floats.
Careful with overflows with integer data types
>>> camera = data.camera()
>>> camera.dtype
dtype('uint8')
>>> camera_multiply = 3 * camera
Different integer sizes are possible: 8-, 16- or 32-bytes, signed or unsigned.
Warning
An important (if questionable) skimage convention: float images
are supposed to lie in [-1, 1] (in order to have comparable contrast for
all float images)
>>> from skimage import img_as_float
>>> camera_float = img_as_float(camera)
>>> camera.max(), camera_float.max()
(255, 1.0)
Some image processing routines need to work with float arrays, and may hence output an array with a different type and the data range from the input array
>>> from skimage import filters
>>> camera_sobel = filters.sobel(camera)
>>> camera_sobel.max()
0.591502...
Utility functions are provided in skimage to convert both the
dtype and the data range, following skimage’s conventions:
util.img_as_float, util.img_as_ubyte, etc.
See the user guide for more details.
3.3.2.2. Colorspaces¶
Color images are of shape (N, M, 3) or (N, M, 4) (when an alpha channel encodes transparency)
>>> face = sp.datasets.face()
>>> face.shape
(768, 1024, 3)
Routines converting between different colorspaces (RGB, HSV, LAB etc.)
are available in skimage.color : color.rgb2hsv, color.lab2rgb,
etc. Check the docstring for the expected dtype (and data range) of input
images.
3.3.3. Image preprocessing / enhancement¶
Goals: denoising, feature (edges) extraction, …
3.3.3.1. Local filters¶
Local filters replace the value of pixels by a function of the values of neighboring pixels. The function can be linear or non-linear.
Neighbourhood: square (choose size), disk, or more complicated structuring element.
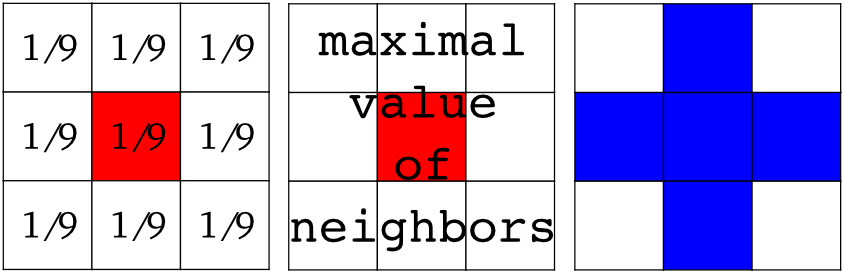
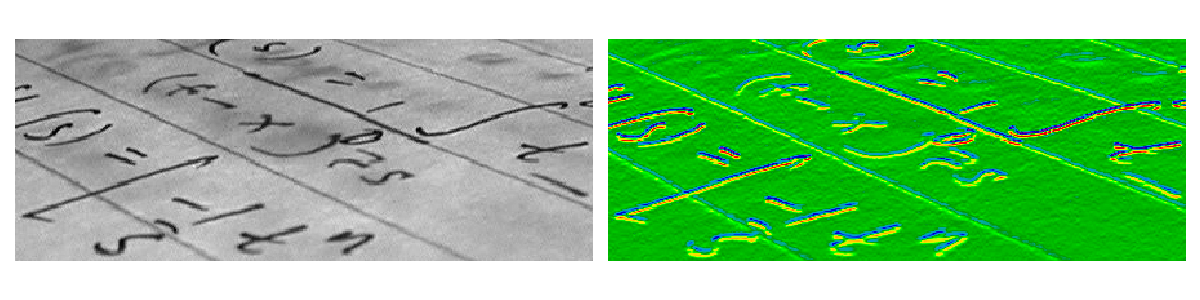
Example : horizontal Sobel filter
>>> text = data.text()
>>> hsobel_text = filters.sobel_h(text)
Uses the following linear kernel for computing horizontal gradients:
1 2 1
0 0 0
-1 -2 -1
3.3.3.2. Non-local filters¶
Non-local filters use a large region of the image (or all the image) to transform the value of one pixel:
>>> from skimage import exposure
>>> camera = data.camera()
>>> camera_equalized = exposure.equalize_hist(camera)
Enhances contrast in large almost uniform regions.

3.3.3.3. Mathematical morphology¶
See wikipedia for an introduction on mathematical morphology.
Probe an image with a simple shape (a structuring element), and modify this image according to how the shape locally fits or misses the image.
Default structuring element: 4-connectivity of a pixel
>>> from skimage import morphology
>>> morphology.diamond(1)
array([[0, 1, 0],
[1, 1, 1],
[0, 1, 0]], dtype=uint8)
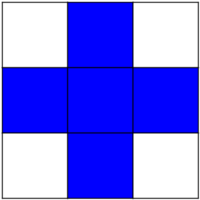
Erosion = minimum filter. Replace the value of a pixel by the minimal value covered by the structuring element.:
>>> a = np.zeros((7,7), dtype=np.uint8)
>>> a[1:6, 2:5] = 1
>>> a
array([[0, 0, 0, 0, 0, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 1, 1, 1, 0, 0],
[0, 0, 0, 0, 0, 0, 0]], dtype=uint8)
>>> morphology.binary_erosion(a, morphology.diamond(1)).astype(np.uint8)
array([[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 1, 0, 0, 0],
[0, 0, 0, 1, 0, 0, 0],
[0, 0, 0, 1, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0]], dtype=uint8)
>>> #Erosion removes objects smaller than the structure
>>> morphology.binary_erosion(a, morphology.diamond(2)).astype(np.uint8)
array([[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0],
[0, 0, 0, 0, 0, 0, 0]], dtype=uint8)
Dilation: maximum filter:
>>> a = np.zeros((5, 5))
>>> a[2, 2] = 1
>>> a
array([[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 1., 0., 0.],
[0., 0., 0., 0., 0.],
[0., 0., 0., 0., 0.]])
>>> morphology.binary_dilation(a, morphology.diamond(1)).astype(np.uint8)
array([[0, 0, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 1, 1, 1, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 0, 0]], dtype=uint8)
Opening: erosion + dilation:
>>> a = np.zeros((5,5), dtype=int)
>>> a[1:4, 1:4] = 1; a[4, 4] = 1
>>> a
array([[0, 0, 0, 0, 0],
[0, 1, 1, 1, 0],
[0, 1, 1, 1, 0],
[0, 1, 1, 1, 0],
[0, 0, 0, 0, 1]])
>>> morphology.binary_opening(a, morphology.diamond(1)).astype(np.uint8)
array([[0, 0, 0, 0, 0],
[0, 0, 1, 0, 0],
[0, 1, 1, 1, 0],
[0, 0, 1, 0, 0],
[0, 0, 0, 0, 0]], dtype=uint8)
Opening removes small objects and smoothes corners.
Higher-level mathematical morphology are available: tophat, skeletonization, etc.
See also
Basic mathematical morphology is also implemented in
scipy.ndimage.morphology. The scipy.ndimage implementation
works on arbitrary-dimensional arrays.
3.3.4. Image segmentation¶
Image segmentation is the attribution of different labels to different regions of the image, for example in order to extract the pixels of an object of interest.
3.3.4.1. Binary segmentation: foreground + background¶
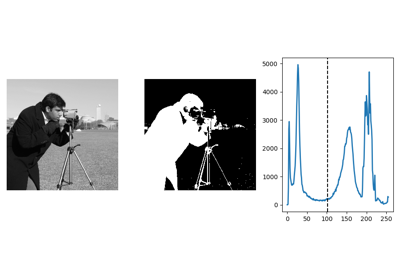
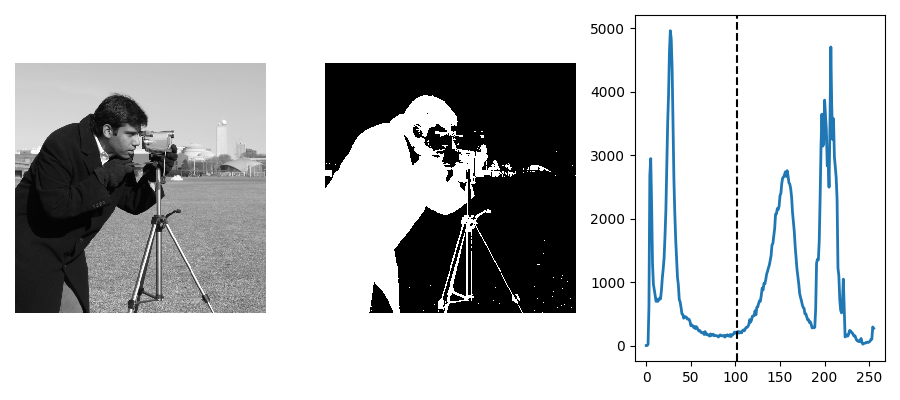
Histogram-based method: Otsu thresholding¶
Tip
The Otsu method is a simple heuristic to find a threshold to separate the foreground from the background.
from skimage import data
from skimage import filters
camera = data.camera()
val = filters.threshold_otsu(camera)
mask = camera < val
Labeling connected components of a discrete image¶
Tip
Once you have separated foreground objects, it is use to separate them from each other. For this, we can assign a different integer labels to each one.
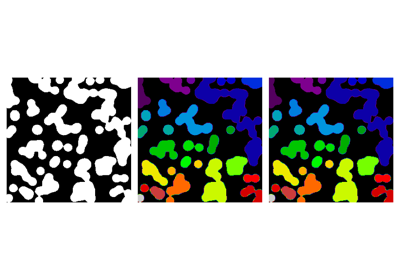
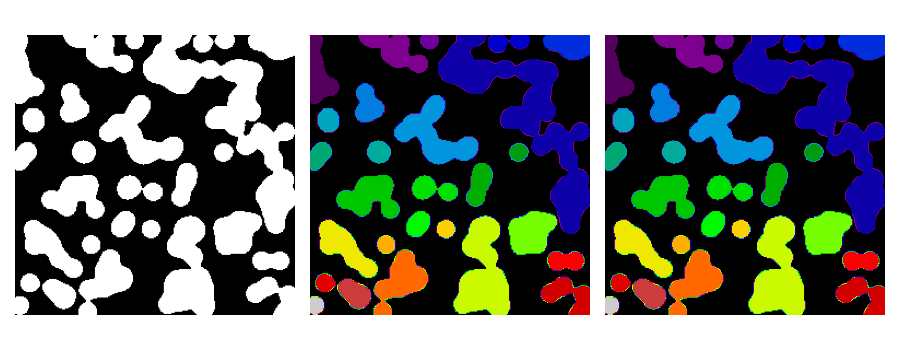
Synthetic data:
>>> n = 20
>>> l = 256
>>> im = np.zeros((l, l))
>>> points = l * np.random.random((2, n ** 2))
>>> im[(points[0]).astype(int), (points[1]).astype(int)] = 1
>>> im = filters.gaussian(im, sigma=l / (4. * n))
>>> blobs = im > im.mean()
Label all connected components:
>>> from skimage import measure
>>> all_labels = measure.label(blobs)
Label only foreground connected components:
>>> blobs_labels = measure.label(blobs, background=0)
See also
scipy.ndimage.find_objects() is useful to return slices on
object in an image.
3.3.4.2. Marker based methods¶
If you have markers inside a set of regions, you can use these to segment the regions.
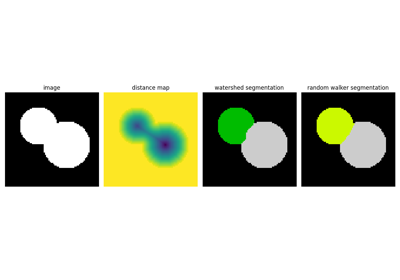
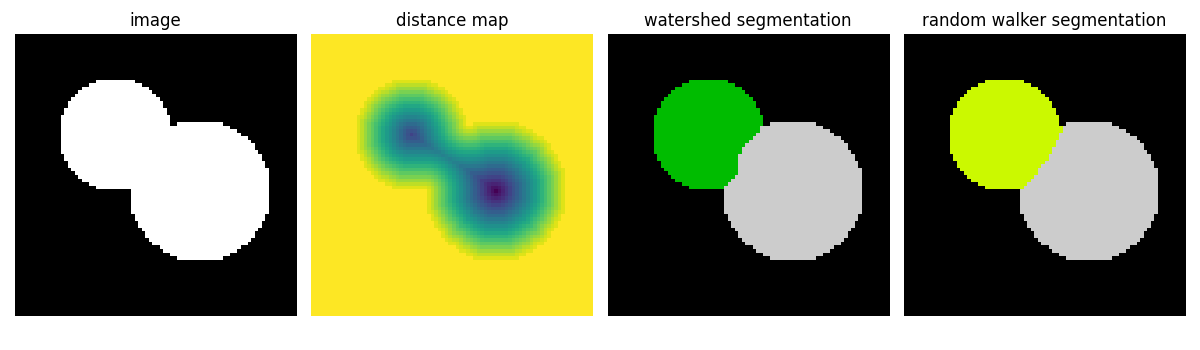
Watershed segmentation¶
The Watershed (skimage.segmentation.watershed()) is a region-growing
approach that fills “basins” in the image
>>> from skimage import morphology
>>> from skimage.segmentation import watershed
>>> from skimage.feature import peak_local_max
>>>
>>> # Generate an initial image with two overlapping circles
>>> x, y = np.indices((80, 80))
>>> x1, y1, x2, y2 = 28, 28, 44, 52
>>> r1, r2 = 16, 20
>>> mask_circle1 = (x - x1) ** 2 + (y - y1) ** 2 < r1 ** 2
>>> mask_circle2 = (x - x2) ** 2 + (y - y2) ** 2 < r2 ** 2
>>> image = np.logical_or(mask_circle1, mask_circle2)
>>> # Now we want to separate the two objects in image
>>> # Generate the markers as local maxima of the distance
>>> # to the background
>>> import scipy as sp
>>> distance = sp.ndimage.distance_transform_edt(image)
>>> peak_idx = peak_local_max(distance, footprint=np.ones((3, 3)), labels=image)
>>> peak_mask = np.zeros_like(distance, dtype=bool)
>>> peak_mask[tuple(peak_idx.T)] = True
>>> markers = morphology.label(peak_mask)
>>> labels_ws = watershed(-distance, markers, mask=image)
Random walker segmentation¶
The random walker algorithm (skimage.segmentation.random_walker())
is similar to the Watershed, but with a more “probabilistic” approach. It
is based on the idea of the diffusion of labels in the image:
>>> from skimage import segmentation
>>> # Transform markers image so that 0-valued pixels are to
>>> # be labelled, and -1-valued pixels represent background
>>> markers[~image] = -1
>>> labels_rw = segmentation.random_walker(image, markers)
3.3.5. Measuring regions’ properties¶
>>> from skimage import measure
Example: compute the size and perimeter of the two segmented regions:
>>> properties = measure.regionprops(labels_rw)
>>> [float(prop.area) for prop in properties]
[770.0, 1168.0]
>>> [prop.perimeter for prop in properties]
[100.91..., 126.81...]
See also
for some properties, functions are available as well in
scipy.ndimage.measurements with a different API (a list is
returned).
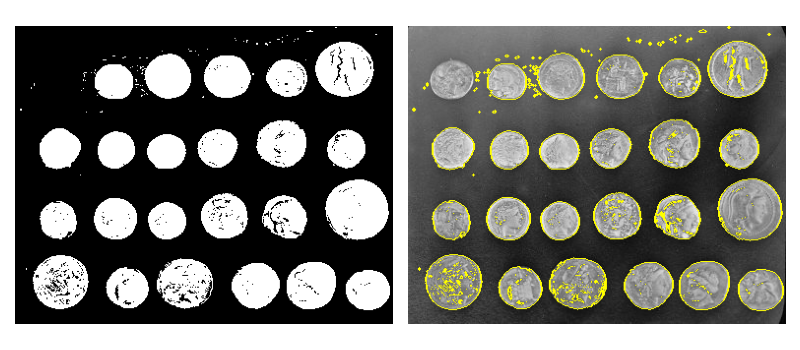
3.3.6. Data visualization and interaction¶
Meaningful visualizations are useful when testing a given processing pipeline.
Some image processing operations:
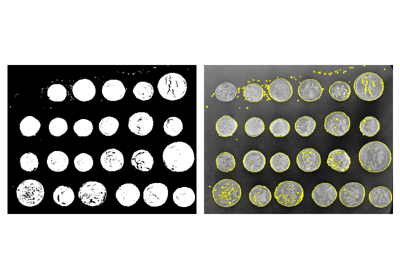
>>> coins = data.coins()
>>> mask = coins > filters.threshold_otsu(coins)
>>> clean_border = segmentation.clear_border(mask)
Visualize binary result:
>>> plt.figure()
<Figure size ... with 0 Axes>
>>> plt.imshow(clean_border, cmap='gray')
<matplotlib.image.AxesImage object at 0x...>
Visualize contour
>>> plt.figure()
<Figure size ... with 0 Axes>
>>> plt.imshow(coins, cmap='gray')
<matplotlib.image.AxesImage object at 0x...>
>>> plt.contour(clean_border, [0.5])
<matplotlib.contour.QuadContourSet ...>
Use skimage dedicated utility function:
>>> coins_edges = segmentation.mark_boundaries(coins, clean_border.astype(int))
3.3.7. Feature extraction for computer vision¶
Geometric or textural descriptor can be extracted from images in order to
classify parts of the image (e.g. sky vs. buildings)
match parts of different images (e.g. for object detection)
and many other applications of Computer Vision
>>> from skimage import feature
Example: detecting corners using Harris detector
from skimage.feature import corner_harris, corner_subpix, corner_peaks
from skimage.transform import warp, AffineTransform
tform = AffineTransform(scale=(1.3, 1.1), rotation=1, shear=0.7,
translation=(210, 50))
image = warp(data.checkerboard(), tform.inverse, output_shape=(350, 350))
coords = corner_peaks(corner_harris(image), min_distance=5)
coords_subpix = corner_subpix(image, coords, window_size=13)
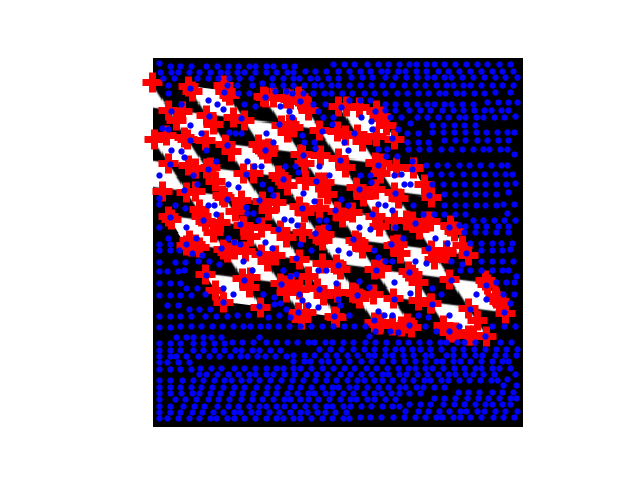
(this example is taken from the plot_corner example in scikit-image)
Points of interest such as corners can then be used to match objects in different images, as described in the plot_matching example of scikit-image.
3.3.8. Full code examples¶
3.3.9. Examples for the scikit-image chapter¶
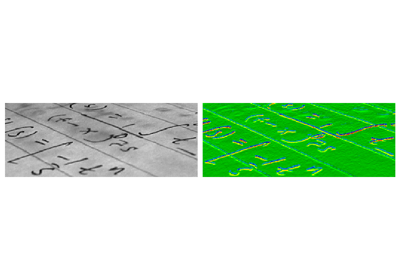
Computing horizontal gradients with the Sobel filter